A course that focuses on topics at the forefront of experimentation in the fields of cellular biology, molecular biology, biochemistry, and genetics.
Learn about Open & Free OLI courses by visiting the “Open & Free features” tab below.
Modern Biology — Open & Free
- Description
- What students will learn
- Learning objectives by module
- Course outline
- Other course details
- System requirements
- Open & Free features
Description
The Modern Biology course covers specialized and somewhat advanced topics in the fields of cellular biology, molecular biology, biochemistry, and genetics. It does not cover organismal biology or taxonomy. The course is carefully planned to provide the background that biology students will need for advanced biology classes. Non-biology majors will also find this course useful as it explains many of the concepts and techniques currently discussed in the popular press.
This course is built around six key concepts that provide unifying explanations for how and why structures are formed and processes occur throughout your study of biology. Because it is not possible to cover the breadth of modern molecular biology in one semester, an understanding of these key concepts will provide a basis for extension of your knowledge to biological systems beyond the specifics covered in this course. One of the major goals of the course therefore is for you to not only learn the definitions of the concepts but also learn to recognize when they are operating the process being studied.
Topics Covered:
- Biological Chemistry
- Cell Biology
- Molecular biology
- Metabolism
Changes in this update include: Improved feedback in many activities, additional interactive 3-D chemical structures, and improved the treatment of metabolism.
What students will learn
Topics Covered:
- Biological Chemistry
- Cell Biology
- Molecular biology
- Metabolism
Learning objectives by module
Unit 2: Biological Chemistry
- Module 2: Atoms, Functional Groups, and Water
- After delving into the functional grouping of molecules, students will be able to identify different functional groups, and characterize each, by category (i.e. polar, non polar, neutral, or charged).
- Define a buffer and its utility to the biological system.
- Define a hydrogen bond and draw possible hydrogen bonds between any two appropriate molecules.
- Define and draw the structure of a hydrogen bond between any two appropriate molecules.
- Define, and draw the structure of, a hydrogen bond between any two appropriate molecules.
- Describe an instance of bioselectivity resulting from the structure and bonding of carbon.
- Describe how water, as a solvent, buffers the structure of ions in solution.
- Describe properties of water that make it the most suitable to support living systems.
- Describe the difference between covalent and non-covalent bonds.
- Describe the energy associated with breaking covalent and non-covalent bonds.
- Describe the hydrophobic effect and its significance in biological systems.
- Describe the properties of water that are critical for the support of living systems.
- Describe the results of the hydrophobic effect, which is the interaction of hydrophobic molecules to the exclusion of water.
- Determine the charge on functional groups at a given a pH level.
- Determine the total number of hydrogen bonds possible from a given electronegative atom.
- Distinguish between a strong electrolyte and a weak electrolyte.
- Explain the basic structure of a water molecule and how it can form a three dimensional structure.
- Explain the significance of having electronegative atoms in a molecule.
- Identify different functional groups and characterize each by category (i.e. polar/nonpolar,charged/uncharged).
- Identify the electronegative atoms found in biological systems.
- Identify the more electronegative atom when comparing two atoms.
- List at least 4 of the major trace elements found in cells and the ionic state in which they exist naturally.
- List atoms (elements) found in biological systems that are most important to life, including the most common atoms and trace elements.
- List the six major atoms found in the composition of biological systems and describe the properties that make them essential for life.
- Recognize and describe a covalent and an ionic bond structurally.
- Recognize and describe a covalent bond and inter-molecular forces (an ionic, a hydrogen bond, and van der Waals interactions) both structurally and energetically.
- Recognize and describe a covalent, an ionic, and a hydrogen bond, both structurally and energetically.
- Module 3: Equilibrium and pH
- Define a weak electrolyte and write an expression for the equilibrium constant.
- Describe the dynamics of equilibrium.
- Describe the relationship between pH and proton concentration.
- Distinguish acids from bases, and explain their relationship to acid dissociation.
- Explain the relationship between the concentration of acid and conjugate base at equilibrium.
- Upon completing this review, students will be able to describethe significance of Ka and pKa, and explain the relationship between them.
- Module 4: Carbohydrates and Polysaccharides
- After examining carbohydrate molecules, students will be able to describe the three major functions of carbohydrates.
- Characterize the functional groups in carbohydrates.
- Define a glycosidic bond and the relationship between condensation and hydrolysis.
- Define the criteria for formation of a glycosidic bond.
- Describe condensation and hydrolysis.
- Describe the anomeric carbons of the hemiacetal form of the carbohydrates.
- Designate functions for the mono-, di-, and polysaccharides, given in the text.
- Distinguish aldose (aldehydes) from ketoses (ketones).
- Distinguish and describe the differences between the plant cell wall and bacterial cell wall.
- Distinguish between an aldose and a ketose.
- Distinguish characteristic structural differences among homopolysaccharides.
- Distinguish primary, secondary, tertiary, and quaternary structures.
- Distinguish, and characterize the differences, between cellulose, starch and glycogen.
- Explain alpha helix and beta structures.
- Explain why all amino acids are the L-isomer and why glycine is not.
- Explain why carbohydrates are well suited for signaling and cell recognition functions.
- Identify a glycosidic bond.
- Identify and describe characteristics of the molecules marked.
- Identify asymmetric (chiral) centers.
- Identify enantiomers of all carbohydrates as D.
- Identify the basic structure of all amino acids.
- Identify the molecules: glyceraldehyde, dihydroxyacetone, ribose, glucose, galactose and fructose.
- Identify which steroisomer of carbohydrates are found in nature.
- Illustrate the different types of bonding responsible for holding the tertiary structure of protein together.
- Place any amino acid into one of the four categories of properties, based on the structure of its side chain.
- Recognize a peptide (amide bond) and list the structural properties of it.
- Recognize alpha and beta orientation of hydroxyl on anomeric carbon.
- Recognize and describe structure and function of sucrose, lactose, cellulose, glycogen, amylose, amylopectin, and starch.
- Recognize the anomeric carbon in the hemiacetal form of the carbohydrates.
- Module 5: Amino Acids and Proteins
- Classify amino acid side chains based on polarity and ionization.
- Define and describe the primary structure of a protein.
- Define the driving force for the folding of a polypeptide in water and in a non-polar solvent.
- Define the structural properties of a peptide bond that will put constraints on the folding of a protein.
- Describe how a quaternary structure is dynamic.
- Describe specific examples of secondary structure in detail.
- Describe specific structural restrictions that characterize secondary structure.
- Describe structural domains.
- Describe the bonding involved in the folding of proteins.
- Describe the type of reaction responsible for the formation anddegradation of the peptide bond.
- Describe what is necessary to form a quaternary structure.
- Explain how primary structure defines the final structure of a protein.
- Explain the covalent bonds possible in stabilizing a tertiary structure.
- Explain why most naturally occurring amino acids are the L-isomers and why glycine is not.
- Give at least one example of a quaternary structure.
- Identify and draw hydrogen bonding between peptide bonds.
- Identify the basic structure of amino acids.
- Identify the functional groups involved in formation of a peptide bond.
- Module 6: Protein Function – Binding and Catalysis
- Define activation energy and the effect an enzyme hasupon it.
- Define equilibrium binding and describe how it is dynamic.
- Describe how pH affects enzyme kinetics.
- Describe how temperature affects enzyme kinetics.
- Describe the effect of changing ligand concentration on an equilibrium.
- Describe the general reaction for an enzyme catalyzed reaction.
- Describe the interactions that stabilize the protein-ligandcomplex.
- Discern the difference between a ligand and a substrate.
- Draw a graph of equilibrium binding.
- Explain the parallels between protein-ligand binding and weak electrolyte dissociation.
- Explain what happens before and after the formation of the ES complex.
- Explain, and graphically illustrate, the effect of each inhibitor type on a velocity vs. substrate concentration graph: a) non-competitive and b) competitive.
- Graphically represent the relationship, between each of the following, for an enzyme catalyzed reaction: 1) velocity and substrate concentration; 2) velocity and enzyme concentration; 3) velocity and pH; and, 4) velocity and temperature.
- Recognize, and describe, the reversible binding of proteins with their ligands.
- Upon completion of this topic, students will be able to recognize, and describe, the reversible binding of proteins with their ligands.
Unit 3: Cell Biology
- Module 7: Lipids and Membranes
- Define the amphipathic character of a phospholipid and glycolipid.
- Define the functions of each of the classes of lipids.
- Describe and diagram the characteristic features of the fluid mosaic membrane.
- Describe and identify the difference between a liposome and a micelle.
- Explain how the different structural features of fatty acids influence their role in phospholipids and fats.
- Name and identify the ester bonds between fatty acids and glycerol and the glycerol and phosphate.
- Module 8: Membrane Transport
- Characterize the environmental changes necessary to allow recycling of intermediates in receptor mediated endocytosis.
- Describe a set of criteria for selective transport of a given molecule to pass through a membrane channel.
- Describe a typical source of energy (general and specific examples).
- Describe how osmotic pressure is generated and what conditions are necessary to create high osmotic pressure in a cell.
- Describe the difference between general endocytosis and protein transduction.
- Describe the factors that affect simple diffusion
- Describe the fate of a molecule taken into a cell by receptor mediate endocytosis.
- Describe the fate of the material undergoing endocytosis.
- Describe the general structural features of a membrane transport protein.
- Describe the solute differences between isotonic, hypertonic and hypotonic solutions.
- Describe the spontaneous direction of the movement of molecules
- Describe the structural and chemical characteristics of a typical ‘facilitator’.
- Describe why the process requires energy.
- Disntiguish how receptor mediated endocytosis differs from protein transduction.
- Distinguish between facilitated diffusion and active transport.
- Distinguish between simple diffusion and facilitated diffusion.
- Distinguish the mechanisms of uniports, symports and antiports.
- Explain how receptor mediated endocytosis differs from phagocytosis.
- Explain the affect of an isotonic, hypertonic and hypotonic solution on the shape of a cell.
- Explain the difference between pinocytosis and phagocytosis.
- Explain why the description of osmosis emphasizes the solvent hanges.
- Expound on how the inside surface of the endocytic vesicle is the same as the outside surface of the cell and why that is important to the cell.
Unit 4: Basis of Molecular Biology
- Module 10: DNA Replication
- Characterize the direction of DNA synthesis.
- Describe one method of editing that takes place during DNA synthesis.
- Describe the formation of the Open Complex at the origin of replication.
- Describe the process of connecting the Okazaki fragments into a continuous strand of DNA.
- Describe the results of semiconservative replication.
- Differentiate the multiple origins of replication that are used to reduce the time of replication of Eukaryotic chromosomes.
- Discuss how competitive binding is used in DNA sequencing using dideoxy-NTPs.
- Distinguish the difference between continuous and discontinuous DNA synthesis.
- Explain how the appropriate substrate for DNA polymerase at the replication fork is established.
- Explain how the the length of DNA amplified during PCR is defined.
- Explain the substrate requirements for DNA synthesis by DNA polymerase.
- Expound on why Okazaki fragments are formed on the lagging strand.
- Specify how telomerase uses the rules for DNA synthesis to overcome shortening of the chromosome.
- Module 11: DNA Transcription
- Describe a post-transcriptional modification that takes place with each of the classes of transcription products.
- Describe the difference between a eukaryotic and prokaryotic Operon
- Describe the differences between the substrate requirements for DNA polymerase and RNA polymerase
- Describe the process of RNA splicing including the splicesome, introns, exons and lariats.
- Describe the role of the 5′-cap and the 3′-polyA tail in the functioning of mRNA.
- Describe the similarities between the functioning of DNA plymerase and RNA polymerase
- Describe the steps in the process of transcription (RNA synthesis) starting with RNA polymerase binding to the promoter and ending with the release of polymerase from the terminator
- Describe where the information (signal) for splicing is and why it might lead to alternative splicing.
- Explain the difference between a weak and a strong promoter including the basis for the difference in “strength” of the promoter
- Name the products of transcription and differentiate each from the others
- Module 12: RNA Translation-Protein Synthesis
- Define the genetic code.
- Define the role of each RNA molecule used in translation.
- Describe the process of translation as a step-wiseprocess.
- Describe what post-transcriptional modification must take place beforetranslation.
- Distinguish between prokaryotic and eukaryotictranslation.
- Module 9: DNA and RNA
- Correctly draw the hydrogen bonding between DNA and RNA bases.
- Correctly identify and specifically name all of the nucleotides.
- Define which helix type describes DNA and RNA.
- Describe 3 common structural characteristics of DNA/RNA polymer backbones.
- Describe the criteria for determining the most stable structure between two DNA molecules, two RNA molecules, between a DNA and a RNA molecule.
- Describe the difference and identify the difference between a nucleotide and a nuceloside.
- Describe the hydrogen bonding that exists between complimentary base pairs in DNA and RNA.
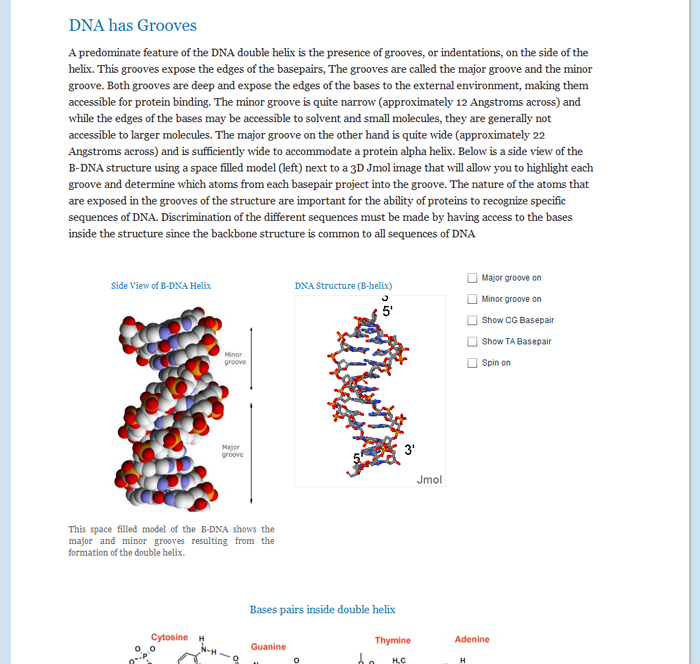
- Describe the major characteristics of the B-DNA double helix
- Describe the structural differences between the A-helix and B-helix.
- Describe why AT and AU base pairs are weaker than GC base pairs.
- Describe why purines are always paired with pyrimidines to form the helix structures of DNA and RNA.
- Explain the major difference between the DNA and RNA backbone structures.
- Give two examples of bioselectivity in the formation of the RNA or DNA backbone.
- Identify a phosphodiester bond.
- Identify correct base pairing between DNA and RNA bases.
- Identify the difference between a purine and pyrimidine.
- Identify the structure of each of the nitrogenous bases: A, T, G, C,U.
- Identify which bases are incorporated into DNA and RNA.
Unit 5: Metabolism
- Module 13: Pathways
- Describe a linear, branched, and circular pathway.
- Describe the common features of metabolic pathways.
- Describe the major differences between a competitive and uncompetitive inhibitor.
- Distinguish regulation by non-covalent and covalent modification of enzymes.
- Explain activation of enzymes by allosteric compounds as a method of regulation.
- State the major differences between catabolic and anabolic pathways.
- Module 14: Energetics
- Calculate the Gibbs free energy change, given the state of the system and the standard energy changes.
- Describe how a pathway with a positive Gibbs free energy can be reversed by coupling to an energy releasing reaction.
- Describe the direction of electron transfer in oxidation/reduction reactions.
- Determine the spontaneous direction of a reaction from the Gibbs free energy.
- Explain how to balance redox reactions.
- Explain the difference between direct and indirect coupling.
- Explain the major energy storage methods in metabolism – phosphorylated compounds, redox carriers, and proton gradient.
- Explain the relationship between the equilibrium point of a system and the difference in standard energies.
- Identify and describe the source of “high-energy” phosphate bonds in ATP.
- Specify the difference between standard energy and Gibbs free energy.
- State the name of two common organic electron carriers and show familiarity with changes in the structure that occurs on oxidation/reduction.
- Tell how the energy stored in a thioester can be used for ATP synthesis or organic addition reactions.
- Module 15: Enzyme Nomenclature
- Describe the reactions catalyzed by dehydrogenases, hydratases, isomerases, and synthetases.
- Explain the difference between a kinase and a phosphatase
- Module 16: Glycolysis
- Define glycolysis and identify its celluar location.
- Describe anaerobic metabolism and how it effects glycolysis.
- Describe how energy released by oxidation is stored.
- Distinguish how direct and indirect coupling are used to make glycolysis spontaneous.
- Explain what happens when glucose is metabolized through glycolysis.
- Module 17: TCA Cycle
- Describe how glycolysis connects to the TCA cycle and where the TCA cycle occurs.
- Describe how intermediates in the TCA cycle are used to synthesize a wide range of compounds.
- Explain where CO2 is released.
- Expound on where, and in what form, the energy released by the TCA cycle is stored.
- Module 18: Electron Transport and Ox. Phos.
- Explain ATP synthase and the effects of allosteric changes during proton translocation.
- Explain the role of the four complexes, coenzyme Q, and cytochrome C in electron transport.
- Explain what happens when electrons transport through complexes I, III, and IV.
- Identify the pathway of the electrons from FADH2 to oxygen resulting in the formation of water.
- Identify the pathway of the electrons from NADH to oxygen resulting in the formation of water.
- Predict whether the free energy stored in the non-equilibrium hydrogen ion gradient is sufficient to sythesize ATP from the transport of 3 protons.
- Module 19: Integrated Metabolism
- Describe how glycolysis and gluconeogenesis are coordinately regulated by sensing energy levels in the cell.
- Describe how select amino acids enter the oxidative pathways.
- Describe indirect coupling in glycogen synthesis and how glycogen synthase is controlled by protein phosphorylation.
- Describe receptor mediated signal transduction and explain the role of G-proteins and adenyl cyclase.
- Explain how glucose can be synthesized from pyruvate.
- Explain how glycogen degradation by glycogen phosphorylase is controlled by protein phosphorylation.
- Explain how hormone controlled phosphorylation of enzymes affect fructose 2,6 phosphate (F-26-P) levels.
- Explain how the disaccharides sucrose and lactose enter glycolysis.
- Explain the physiological role of the hormones epinephrine, glucagon, and insulin.
- Explain the role of the protein kinases and phosphatases in regulating the enzyme function.
- Express how glycogen and glucose metabolism in the liver effects the glucose demand of the organism and the liver cell.
- Identify the role of phosphofructose kinase and fructose-1,6-bisphosphatase in gluconeogenesis pathways.
- Indicate the number of steps in glycolysis that are too energetically unfavorable to reverse in gluconeogenesis and describe how the steps are reversed.
- Recognize the structure of glycogen.
- Specify how fats (triglycerides) are metabolized by fatty acid oxidation.
- Specify the number of steps in glycolysis with Gibbs free energies that are close to zero and can be reversed in gluconeogenesis.
- Summarize how fructose 2,6, phosphate levels regulate glycolysis and gluconeogenesis.
- Tell how carbons atoms from the excess consumption of any dietary source can be incorporated into fats.
Course outline
UNIT 1: Introduction to Modern Biology
Module 1: Introduction
UNIT 2: Biological Chemistry
Module 2: Atoms, Functional Groups, and Water
Module 3: Equilibrium and pH
Module 4: Carbohydrates and Polysaccharides
Module 5: Amino Acids and Proteins
Module 6: Protein Function – Binding and Catalysis
UNIT 3: Cell Biology
Module 7: Lipids and Membranes
Module 8: Membrane Transport
UNIT 4: Basis of Molecular Biology
Module 9: DNA and RNA
Module 10: DNA Replication
Module 11: DNA Transcription
Module 12: RNA Translation-Protein Synthesis
UNIT 5: Metabolism
Module 13: Pathways
Module 14: Energetics
Module 15: Enzyme Nomenclature
Module 16: Glycolysis
Module 17: TCA Cycle
Module 18: Electron Transport and Ox. Phos.
Module 19: Integrated Metabolism
UNIT 6: Appendix
Module 20: Appendix
Other course details
System requirements
OLI system requirements, regardless of course:
- internet access
- an operating system that supports the latest browser update
- the latest browser update (Chrome recommended; Firefox, Safari supported; Edge and Internet Explorer are supported but not recommended)
- pop-ups enabled
- cookies enabled
Some courses include exercises with exceptions to these requirements, such as technology that cannot be used on mobile devices.
This course’s system requirements:
Open & Free features
Open & Free Courses
- Open & Free OLI courses enable independent learners to study a subject on their own terms, at their leisure. Courses are:
- Self-guided.
- Self-paced.
- Self-supported.
- Open & Free courses include only the learning materials:
- No teacher.
- No tests.
- No college credit.
- No certificate of completion.
- *If your teacher gave you a Course Key, do not use an Open & Free course because your teacher will never see your work.